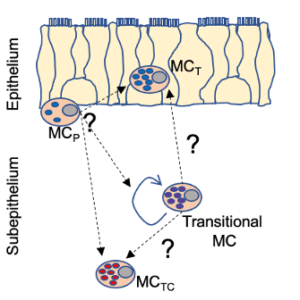
 Mast cells begin their lives as agranular, circulating progenitors in the bone marrow. They are recruited to sites of inflammation throughout the body, after which they integrate signals from their tissue microenvironment and the local inflammatory milieux to take on one of several discrete phenotypes. Our studies in human nasal polyposis and eosinophilic esophagitis additionally suggest that local proliferation also contributes to mast cell expansion in type 2 inflammatory disease, and that progenitors may progress through a plastic “transitional” stage before their final maturation. Little is currently known about either the signals that direct mast cell progenitor recruitment and differentiation, what drives this local mast cell proliferation, or how these different mast cell phenotypes can differentially influence host tissue.
Mast cells begin their lives as agranular, circulating progenitors in the bone marrow. They are recruited to sites of inflammation throughout the body, after which they integrate signals from their tissue microenvironment and the local inflammatory milieux to take on one of several discrete phenotypes. Our studies in human nasal polyposis and eosinophilic esophagitis additionally suggest that local proliferation also contributes to mast cell expansion in type 2 inflammatory disease, and that progenitors may progress through a plastic “transitional” stage before their final maturation. Little is currently known about either the signals that direct mast cell progenitor recruitment and differentiation, what drives this local mast cell proliferation, or how these different mast cell phenotypes can differentially influence host tissue.
My laboratory focuses on understanding how mast cells integrate tissue stroma-derived signals cells to take on the discrete mast cell effector phenotypes found within epithelial and subepithelial regions of mucosal tissues, determining how these effector phenotypes change during inflammation and disease, and defining the relative contributions of each phenotype to disease progression. We’ve developed a series of complementary approaches in human and mouse to address these questions. Using single-cell RNA-sequencing, we define the heterogeneous mature mast cell phenotypes found in human disease and map the progression from circulating progenitor to mature mast cell. Through in-vitro modeling and bulk RNA-seq, we test the influence of individual structural cells on mast cell development determine the “fingerprints” of candidate cytokines and growth factors on the mast cell transcriptome. We then can compare our in vitro results to human mast cell transcriptomes in vivo and test the role of specific candidates on mast cell development and function through mouse modeling, taking advantage of several cre-recombinase expressing strains that allow us to specifically target discrete mast cell lineages.
